All published articles of this journal are available on ScienceDirect.
Role of Protein Tyrosine Phosphatase (PTPN22) Gene [C1858T] Functional Variant in Genetic Susceptibility of Psoriatic Arthritis in Kuwaiti Arabs
Abstract
Background:
Psoriatic arthritis (PsA) is a chronic, systemic inflammatory arthritic disease characterized by joint inflammation that is associated with cutaneous psoriasis, and can lead to pain, swelling, or stiffness in one or more joints. It results from a complex interplay between genetic, immunologic and environmental factors. A functional variant [C1858T] in the protein tyrosine phosphatase (PTPN22) gene, which encoded Arg620Trp in the lymphoid protein tyrosine phosphatase (LYP) has been shown to be a negative regulator of T-cell activation.
Objective:
The objective of this study was to investigate an association between PTPN22 gene [C1858T] functional variant and PsA in Kuwaiti patients.
Methods:
We have investigated the association of PTPN22 gene functional variant in 102 Kuwaiti patients with psoriatic arthritis and compared it to that in 214 healthy controls. The genotypes for the PTPN22 gene [C1858T] variant were determined by using a PCR-RFLP method and confirmed by DNA sequence analysis.
Results:
The frequency of homozygous variant genotype (TT) was found to be significantly higher in PsA patients compared to that in the controls (p <0.0001). Collectively, the variant genotype was detected in homozygous and heterozygous combinations in 30% patients (p <0.0001) compared to 16% in the controls. The frequency of variant genotype was found to be highest in the early-onset PsA patients (age >25-34y). No correlation was detected between the variant genotype (TT) and gender in the Kuwaiti PsA patients.
Conclusion:
Our data show a significant association of PTPN22 gene functional variant [C1958T] with PsA in Kuwaiti patients and highlight its role in determining the genetic susceptibility along with other factors.
1. INTRODUCTION
The autoimmune diseases are characterized by abnormal immune responses against self-tissues and organs that are subjected to continuous inflammation, thus resulting in their damage. Along with the abnormal immune system, genetic predisposition and environmental factors have also been implicated in autoimmune disease pathogenesis [1]. Psoriatic arthritis (PsA) is heterogeneous inflammatory arthritis, which occurs in patients with psoriasis and can cause pain, swelling, or stiffness in joints and may also result in movement disorders [2, 3]. The disease severity has been shown to vary between patients and also within a specific patient over a period of time. The disease manifestation can vary from mild mono-oligoarthritis to severe erosive polyarthritis comparable with Rheumatic Arthritis (RA) [4]. In contrast to RA, manifestations such as dactylitis and enthesitis are common in patients with PsA, as is the case in patients suffering from other diseases within the seronegative spondylarthropathy group [5, 6]. In addition, in contrast to RA, the majority of the individuals with PsA are seronegative for Rheumatoid Factor (RF) and anti-citrullinated protein-peptide antibodies (ACPA) [7, 8]. A number of genes have been associated with PsA in previous reports mainly from Caucasians and some of these genes are common to a number of other autoimmune diseases [9-11]. Epidemiological evidence has demonstrated a high heritability of psoriatic arthritis compared with psoriasis vulgaris [9, 12]. It has been reported that genetic susceptibility can be considered as a contributing factor in approximately 50% PsA patients who have a family history of spondyloarthropathies or cutaneous psoriasis [3]. More recently, the genome-wide association studies (GWAS) have demonstrated that the genetic contributions of psoriatic disease account for less than 25% of heritability [1, 10].
The protein tyrosine phosphatase non-receptor 22 (PTPN22) gene, located on chromosome 1p13, codes for a protein, LYP, which is considered as a negative regulator of T cells [13]. A Single Nucleotide Polymorphism (SNP) rs2476601 [C1858T], located in exon 14 of the PTPN22 gene, results in a functional variant, Arg620Trp, which has been associated with several autoimmune diseases [13-16]. Previous studies investigating an association between PTPN22 gene [C1858T] functional variant and susceptibility to PsA have shown conflicting results [12, 14-16]. These inconsistent results have been attributed to various causes such as small sample sizes, racial or ethnic differences and clinical or genetic heterogeneity [14-16]. Therefore, it is important to study the ethnicity-specific association of PTPN22 gene [C1858T] functional variant with PsA to determine its role in genetic predisposition in different populations.
The aim of this study was to investigate the possible association of PTPN22 gene [C1858T] functional variant in a completely different population/ethnic group (Kuwaiti Arab patients with PsA) and controls in order to determine its impact on genetic susceptibility of PsA.
2. MATERIALS AND METHODS
This study included 102 Kuwaiti Arab patients with psoriatic arthritis (PsA) and 214 healthy controls of matching ethnicity. The PsA patients were seen regularly at the Rheumatic Disease Unit, Amiri Hospital, Kuwait. The diagnosis of PsA was based on the presence of inflammatory arthritis associated with psoriasis, usually with no rheumatoid factor in the serum [5]. The diagnosis of PsA had been made at least six months prior to enrollment in the study. Detailed clinical information was obtained from all patients, including age, gender, age at disease onset, family history of psoriasis and/or PsA, and clinical manifestations. Patients were considered to have ‘early-onset’ PsA if disease onset was at any age younger than 40 years, and ‘late-onset’ PsA if age at onset was after their 40th birthday. The controls were healthy Kuwaiti nationals and were evaluated by a trained Rheumatologist for their health status. They did not have a history of autoimmune or rheumatic disorders or other diseases of known genetic or hereditary predisposition. The subjects included in the study, both patients and controls were unrelated to each other but were matched for age and gender.
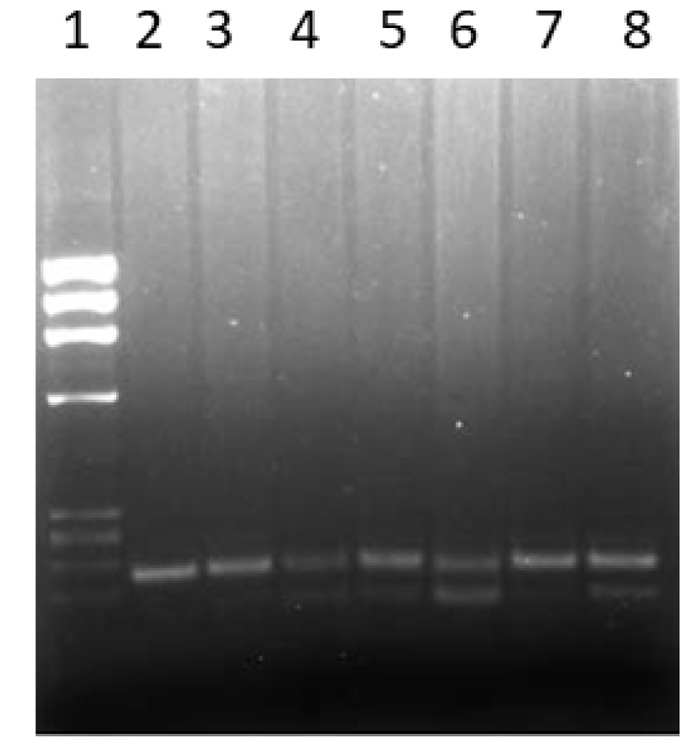
Blood (approximately 5 ml) was withdrawn from all the study subjects and anticoagulated by EDTA. A previously described method was used to isolate total genomic DNA [17]. The genotypes of a functional variant +1858C→T (rs2476601) in the PTPN22 gene were identified by polymerase chain reaction-restriction enzyme fragment length polymorphism (PCR-RFLP) method [18, 19]. In order to amplify a 218 bp DNA fragment, the primers: Forward primer: 5’-ACTGATAATGTTGCTTCAACGG-3’ and reverse primer:5’-TCACCAGCTTCCTCAACCAC-3’ were used. The PCR mixture contained 10x PCR buffer (Applied BioSystems, USA); 0.2 mM of dNTPs (deoxyribonucleotide triphosphates); 1.5 mM MgCl2; 20 pmol of each primer, 200 ng template DNA and 1U AmpliTaq DNA polymerase (Applied BioSystems). The PCR program consisted of denaturation at 940C for 2 min followed by 35 cycles of 940C for 30 seconds, 600C for 30 seconds and 720C for 30 seconds and an extension step at 720C for 7 minutes. The amplification products were digested with restriction enzyme RsaI at 370C for 90 min. The cleavage products were analyzed by 2% agarose gel electrophoresis and visualized under UV light after staining with ethidium bromide. The absence of RsaI restriction enzyme site in the 218 bp PCR product was associated with the 1858T allele, while in the presence of 1858C allele, 176 bp and 42 bp cleavage products were detected. PCR products from a heterozygous individual produced the 218, 176 and 42 bp cleavage products. Sanger’s DNA sequencing method was used to sequence the PCR amplicons on ABI 3130 genetic analyzer to confirm the genotypes.
The data collected were analyzed using the Statistical Package for Social Sciences (SPSS) ver.25, Chicago, IL, USA. The frequency of genotypes and alleles from PsA patients and controls were determined by direct counting. The Confidence Interval (CI) was fixed at 95% and statistical significance was set at P <0.05 (two-tailed). Fisher’s Exact test was used to determine the statistical significance of the differences between genotype and allele frequency in PsA patients and controls. For calculation of the statistical significance in co-dominant and dominant models, the genotype frequency in homozygous CC subjects and the ‘C’ allele frequency were considered as reference (assumed to be associated with the least risk of PsA). In the case of the dominant model, the genotype frequencies of CT and TT were combined (PsA patients having at least one variant ‘T’ allele of the PTPN22 gene). One possible limitation of this study might be that in some comparisons, e.g., with age and gender, the presence of low numbers in the study groups could potentially result in type-II errors. The genotype distribution was tested for Hardy Weinberg equilibrium by the goodness of fit method using MSTAT software.
3. RESULTS
The genotypes of the PTPN22 gene functional variant [C1858T] were determined by using the PCR-RFLP method as described in the Methods section. A representative gel showing the method used for the detection of various genotypes of the PTPN22 gene [C1858T] functional variant is presented in Fig. (1). The baseline characteristics of Kuwaiti PsA patients included in this study have been presented in Table 1.
The homozygous variant (TT) genotype frequency of the PTPN22 gene C1858T variant was found to be significantly higher in Kuwaiti PsA patients than that in the controls (OR 9.06, Table 2). Collectively, the homozygous (TT) and heterozygous (CT) genotypes of the PTPN22 gene C1858T polymorphism (i.e., individuals with at least one variant T-allele) were detected in 30% PsA patients compared to 16% in the controls (Table 2). The genotype frequency was not significantly different in the case of heterozygous CT genotype (Table 2). The power calculation for the variant TT genotype (OR 9.06) provided the study power of 100% for the estimation of the PsA risk. The differences were also significant between patients and controls when the allele frequencies of ‘C’ and ‘T’ alleles were compared (P <0.0001; Table 2).
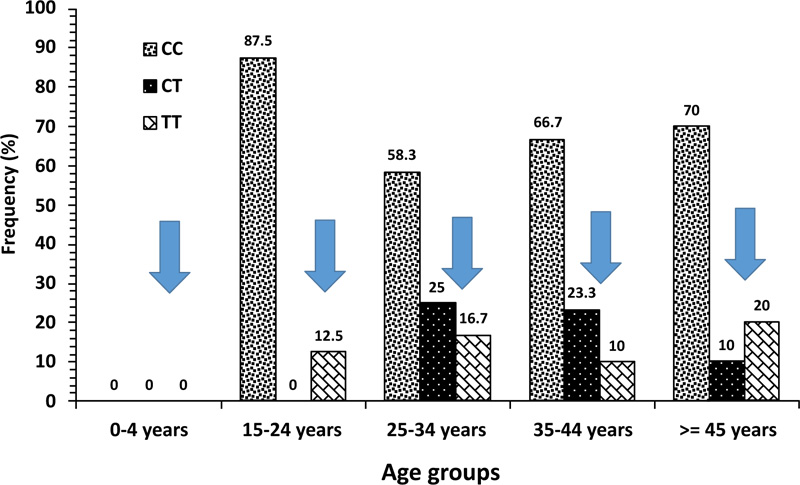
The distribution of PTPN22 gene polymorphism genotypes was compared in Kuwaiti PsA patients stratified on the basis of age-distribution pattern and the data is presented in Fig. (2). The frequency of variant genotypes TT was 0% in 0-4y age group, 12.5% in 15-24y, 16.7% in 25-34y, 10% in 35-44y and 20% in PsA patients with >45y of age (Fig. 2). When the two genotypes, i.e., TT and CT (with at least one variant allele) were considered together, the frequency was 12.5% in 15-24y, 42% in 25-34y, 33% in 35-44y and 30% in respective age-groups of Kuwaiti PsA patients (Fig. 2).
| Mean age at diagnosis (+SD) years | 39.2 (+9.0) |
| Median disease duration (range) months | 60 (6-240) |
| Clinical manifestations: | n (%) |
| Asymmetric polyarthritis | 78 (76.4) |
| Symmetric polyarthritis | 14 (13.7) |
| Oligo-arthritis | 10 (9.8) |
| Spondylitis | 20 (19.6) |
| Dactylitis | 48 (47.1) |
| Enthesitis | 20 (19.6) |
| Genotypes |
Patients N=102 (%) |
Controls N=214 (%) |
OR (95% CI)* | P-value** |
|---|---|---|---|---|
| Co-dominant | ||||
| CC | 71 (70) | 180 (84) | 1.0 (Reference)*** | |
| CT | 15 (14) | 32 (15) | 1.19 (0.61 – 2.33) | 0.6 |
| TT | 16 (16) | 2 (1) | 9.06 (2.55 – 14.50) | <0.0001 |
| Dominant | ||||
| CC | 71 (70) | 180 (84) | 1.0 (Reference) | |
| CT/TT | 31 (30) | 34 (16) | 2.31 (1.32 – 4.04) | 0.005 |
| Alleles | N = 204 (%) | N = 428 (%) | ||
| C-allele | 157 (77) | 392 (92) | 1.0 (Reference) | |
| T-allele | 47 (23) | 36 (8) | 3.26 (2.03 – 5.23) | <0.0001 |
| PTPN22 genotype/ gender | Patients n (%) | Control n (%) | OR (95% CI)* | P-value** |
|---|---|---|---|---|
| n=71 | n=177 | |||
| Male CC | 33 (46.5) | 62 (35) | 1.61 (0.92 – 2.82 | 0.13 |
| Female CC | 38 (53.5) | 115 (65) | 0.62 (0.35 – 1.09) | 0.13 |
| n=15 | n=32 | |||
| Male CT | 3 (20) | 13 (40.6) | 0.37 (0.09 – 1.57) | 0.20 |
| Female CT | 12 (80) | 19 (59.4) | 2.73 (0.64 – 8.66) | 0.20 |
| n=16 | n=2 | |||
| Male TT | 5 (31.25) | 1 (50) | 0.45 (0.02 – 8.84) | 1.00 |
| Female TT | 11 (68.75) | 1 (50) | 2.20 (0.11 – 9.76) | 1.00 |
The genotype frequencies of PTPN22 gene functional variant [C1858T] were compared between patients and controls in relation to the gender and the data is presented in Table 3. No correlation was found between gender and the manifestation of PsA in Kuwaiti patients (Table 3). In the case, of PsA patients with TT genotype, majority 11/16 (69%) were females, while in comparison, TT genotype was detected in 31% of the male PsA patients (Table 3). A similar distribution pattern was noted in the case of heterozygous CT genotype, in which case, 80% of the females had the CT genotype (Table 3).
4. DISCUSSION
The most striking finding in this study is the significant association detected between the homozygous functional variant (TT) genotype of the PTPN22 gene and clinical presentation of psoriatic arthritis in Kuwaiti Arabs. The PTPN22 gene encodes a functional protein tyrosine phosphatase, LYP, which acts as a regulator of the negative regulatory kinase in T cells. It has been shown to play a role in suppressing T cell activation and, therefore, maybe the mechanism by which association is likely to contribute to the genetic susceptibility of PsA and indeed for several other autoimmune disorders [13].
Previous association studies on the PTPN22 gene [C1858T], functional variant and PsA have yielded conflicting results. Hinks et al. [14] did not find an association of this variant with PsA in a patient population from the UK. Similarly, no association was detected between PTPN22 gene [C1858T] functional variant and PsA in German patients when the whole of the patient group was considered; however, in the males, a significantly higher proportion carried the risk allele ‘T’ [15]. A recent study from Greece did not find an association between PTPN22 gene [C1858T] variant and PsA [20]. However, this report from Greece did find an association between TYK2 (tyrosine kinase-2) and STAT4 (signal transducer and activation of transcription-4) genes and PsA and also reported an overlap between PsA and rheumatoid arthritis (RA). In contrast to these negative associations, our results from Kuwaiti Arabs show a significant positive association between PsA and PTPN22 gene [1858T] functional variant (Table 2). However, unlike the study from Greece, we did not find any significant overlap between PsA and RA in the Kuwaiti Arab patients with PsA. In a meta-analysis, which evaluated the risk of psoriasis in relation to PTPN22 gene [C1858T] functional variant, a positive association between psoriasis and the PTPN22 gene 1858T allele was reported and this association was found to be stronger among the subjects with PsA [21]. Butt et al. [16] investigated the association between PTPN22 gene [1858T] functional variant and psoriatic arthritis in two populations. They reported a moderate association of the PTPN22 gene [C1858T] variant with PsA in a patient population from Toronto, but not in another population from Newfoundland, in which no difference in the risk allele (T) frequency was detected [16]. On the basis of these divergent results, the authors argued that the association might be disease and population-specific [16]. A positive association between PTPN22 gene functional variant [C1858T] and PsA has been reported in patients from Sweden [21]. This report also showed that carriers of the variant ‘T’ allele had a significantly higher number of deformed joints [21]. In a study from Saudi Arabia, a weak association between the heterozygous CT genotype of the PTPN22 gene functional variant was detected in psoriasis patients [22]. Interestingly, the homozygous TT genotype was not detected at all in either patients or the controls in this study from Saudi Arabia [22]. Although psoriasis and psoriatic arthritis are interrelated disorders, PsA is a distinct entity with its own epidemiological, clinical and genetic features [1]. Furthermore, PsA exhibits much greater heritability among the first degree relatives than psoriasis [10]. To our knowledge, the majority of genetic association studies on PTPN22 gene with PsA have been carried out in Caucasians and the only study from the Middle East is the one mentioned above from Saudi Arabia on psoriasis [22]. The prevalence of PTPN22 [C1858T] functional variant has been shown to vary in different ethnic groups, e.g., it was found to be low in Asians and Africans and was virtually absent in Han Chinese [23, 24]. It has been reported that in Europe, a distinct North-South gradient exists in the prevalence of the PTPN22 gene [C1858T] functional variant [25].
In our study, the highest frequency of PTPN22 gene functional variant [1858T] was noted in the age range 25-34y amongst the Kuwaiti PsA patients (42% of the PsA patients had at least one variant ‘T’ allele, i.e., combined TT and CT genotypes) compared to 33% in 35-44y age-group and 30% in >45y age-group, respectively (Fig. 2). This demonstrated that a higher relative proportion of early-onset PsA patients (<40y of age) carried at least one ‘T’ risk-allele of the PTPN22 gene functional variant [C1858T], further highlighting its role and contribution in genetic susceptibility of PsA in Kuwaiti Arabs.
We did not find any correlation between the frequency of PTPN22 gene functional variant and the gender in Kuwaiti Arabs with PsA (Table 3). The gender-distribution in our patients showed the presence of a higher number of females in our study group. In the study from Saudi Arabia on psoriasis, it was reported that allele T and CT genotype were more prevalent in female psoriasis patients than in the males, although the differences were not found to be statistically significant [22]. In another study from Germany, the male predominant correlation of variant ‘T’ allele has been reported in PsA patients [15]. However, our results in Kuwaiti PsA patients did not find a gender-correlation.
Kuwaiti Arabs constitute nearly 45% of the population of the State of Kuwait and the population itself is quite diverse. It is a small country located in the north of the Arabian Gulf. There is a high incidence of consanguinity (54%), which has resulted in the familial clustering of chronic disorders [26]. The original settlers of Kuwait had migrated from Najd, an area, which is now part of eastern and central Saudi Arabia. It has been reported that the ethnic origin of Kuwaiti Arabs is diverse; nearly half are of Arab origin, some are Bedouins and the rest are migrants [27]. The Arab citizens of the Gulf States, in general, are thought to have resulted from admixtures between various ethnic groups such as Persians, Turks, South Asians, Europeans and Africans [27]. The genotype frequency of PTPN22 gene function variant [C1858T] reported in this study in Kuwaiti PsA patients is amongst the highest (TT, 16% and combined TT and CT in 30% PsA patients) compared to any other population/ethnic group. This, in our opinion, can possibly be due to a combined effect of unique ethnic/genetic background along with a very high rate of consanguinity in Kuwaiti Arabs and can, therefore, at least partially explain the high incidence of autoimmune diseases, including PsA in Kuwait.
CONCLUSION
Our data demonstrate a significant association of PTPN22 gene [C1858T] functional variant with psoriatic arthritis in Kuwaiti Arabs and highlight its involvement in genetic susceptibility along with other factors. The highest frequency of the homozygous TT genotype of the PTPN22 gene [C1858T] functional variant was detected in the early-onset age-group (25-34y) in Kuwaiti PsA patients, further highlighting its role in genetic susceptibility to PsA.
LIST OF ABBREVIATIONS
| PTPN22 | = Protein Tyrosine Phosphatase Receptor Type N22 |
| PsA | = Psoriatic Arthritis |
| LYP | = Lymphoid Protein Tyrosine Phosphatase |
| PCR | = Polymerase Chain Reaction |
| RFLP | = Restriction Fragment Length Polymorphism |
| RA | = Rheumatoid Arthritis |
| RF | = Rheumatoid Factor |
| ACPA | = Anti-citrullinated Protein-Peptide Antibody |
| GWAS | = Genome-wide Association Study |
| SNP | = Single Nucleotide Polymorphism |
| UV | = Ultra-Violet |
| BP | = Base Pair |
| CI | = Confidence Interval |
| OR | = Odds Ratio |
| STAT4 | = Signal Transduction and Activation of Transcription-4. |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The Ethics Committee of Health Sciences Centre, Kuwait University approved this study (Ref. No. VDR/EC/3243).
HUMAN AND ANIMAL RIGHTS
No Animals were used in this research. All human research procedures followed were in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
CONSENT FOR PUBLICATION
Written informed consent was obtained from all the patients when they were enrolled.
AVAILABILITY OF DATA AND MATERIALS
The authors confirm that the data supporting the findings of this study are available within the article.
FUNDING
This study was funded by the Kuwait University, General Research Support Facility under grant no. GF/Med/03-20.
CONFLICT OF INTERESTS
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We would like to thank the patients for participation in the study and the hospital staff for their assistance.


